New warheads on the way!
NEQUIMED/IQSC/USP, in partnership with groups specialized in parasitology of Trypanosoma cruzi, the agent that causes Chagas disease, identifies new drug candidates for the treatment of this disease. One of the key steps in the drug pipeline at this stage of the process is the scaling-up of the synthesis for the production of the desired grams-scale products. In vivo assays are now essential for the evaluation of potency and early toxicity, with consequent determination of dosage. Synthetic chemists eager to contribute to synthetic medical chemistry with scaled-up and improved reaction yields are welcome to join our group. Interested parties should send a message through our contacts.
The chemical space synthesis
It is not enough to do indiscriminate synthesis of new chemicals to find new drugs. For example, at the time of combinatorial chemistry, a myriad of chemicals could (and can) be synthesized. However, most of the time, without the necessary drug-like characteristics. The main reason for this is the lack of properties adjusted to the desired chemical space such as molecular mass (MM), lipophilia (as logP) and polar surface area (PSA). An extra problem concerns the search for substances free from stereogenic centers which, if neglected, may well lose the natural cunning of the L world. Of course, there are recognized exceptions.
Considering that chemical space can be the universe (see here), it is better to think appropriately how to construct it using, for example, our strategy in molecular planning based on hypotheses (see here and here). In our hypotheses, however, there are some difficult syntheses to do. Perhaps, however, that is why we are taking off to a space of success. Stairway to success is leaning and requires an efficient cycle of plan-synthesize-test-analyze-plan! Repeat the cycle. Then, subnanomolar inhibitors of cysteine proteases appear and some of them kill Trypanosoma cruzi.
Although we are focused on the druggable chemical space, we are attentive to its neighbors.
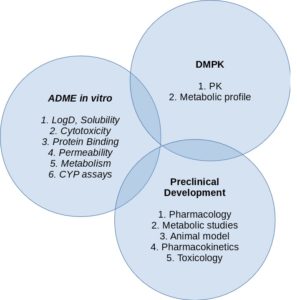
Drug Discovery and Development
Growing Knowledge Expertise
Data Science comes to NEQUIMED/IQSC/USP
The NEQUIMED/IQSC/USP implements an early vocation initiated by one of its members (A neural network analysis for antileishmaniasis compounds). A new strategy that can help in the process of discovering new drug candidates: Machine Learning in systems that mimic the human brain and allow access to huge amounts of data.
It is an innovative computational strategy that quickly and efficiently “teaches” computers to find new, successful bioactive chemicals in a way that supersedes current computer-based methods.
Machine learning encompasses many types of algorithms-including decision trees, nearest neighbors (but mostly see k-medoids), and neural networks that “learn” from training datasets and then make predictions of the real world using test and validation collections. Deep learning is a sophisticated type of multi-layered neural network that optimizes responses. These layers are composed of nodes (which mimic the neurons of the human brain) and are fired in the presence of stimuli.
Along with neural networks, NEQUIMED/IQSC/USP also employs two other techniques of machine learning, namely random forests (RF) and support vector machine (SVM).
Taken together, the high level of abstraction of the data has allowed to identify new inhibitors of cysteine proteases of interest for different disease models that use these proteins enzymes as markers.